


The MarlovitsLab
@ Kulcar/Marlovits
Molecular machines
and targeted therapeutics
@ IMP/IMBA


Design and Structural Plasticity
Function and Disease

——most recent——
ERC synergy - Nov-5th, 2024
.... Delighted and honored to be recepient of an ERC synergy grant (6 years) together with fantastic colleagues - Edith Houben (Amsterdam), Tracy Palmer (Newcastle), John McKinney (Lausanne). Many thanks to panel members, reviewers, colleagues, lab members and supporting institutions.
Exciting science ahead!
PostDoc and PhD positions within a unique and fun international consortium. Get in touch!
Science Advances (2023)
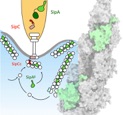
.... Novel coordinated molecular strategy employed by two bacterial proteins originating from the T3SS (Salmonella) to effectively hijack eukaryotic cells. Both, SipC and SipA, work on actin to subvert the host cell cytoskeleton locally at the site of infection. SipA binds in a unique pattern and stabilizes young actin under substoichiometric conditions. Fantastic work by Biao, Rory and collaborators Irina&Sabine (UKE), Jonas&Jan (et al MHH) and Roland (CSSB,UHH,LIV).
doi.org10.1126/sciadv.adj5777
Cell (2023)
.... Discovery of highly neutralizing human antibodies targeting Pseudomonas aeruginosa. Some CFTR patients are resistant to Pseudomonas infections. Here, we identified the binding epitope of neutralizing human antibodies targeting PcrV, the tip protein of the T3SS. Great collaboration with Alex and Jan et al. Fantastic cryoEM by Biao.
doi.org10.1016/j.cell.2023.10.002
mSphere (2023)
.... Discovery of PPE proteins as outer membrane transport proteins for T7SS. Great collaboration with Edith and Wilbert. Mastered by Catalin.
doi.org10.1128/msphere.00402-23
Current Opinion in Structural Biology (2023)
.... Review about the AAA+ ATPases in RuvAB-HJ. Thanks Jirka!
doi.org10.1016/j.sbi.2023.102650
J Struct Biol (2023) - PickYOLO
.... Ultra-fast particle annotator for cryo electorn tomograms. May allow on-the-fly particle detection and subtomogram image data processing. Collaboration with Philipp and his team. Try it out!
doi.org/10.1016/j.jsb.2023.107990
Structure (2023) - MEDIC built in StarMap
.... Tool to detect model errors in atomic models built into ‘density maps’. The function is built into StarMap found in ChimeraX’s toolshed. Great collaboration with Frank and his team. Try it out!
doi.org/doi: 10.1016/j.str.2023.05.002
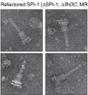
Nature Protocols (2022) - StarMap
.... Tool to easily set-up molecular refinements into cryoEM maps and analyze structural models.
Combines Monte Carlo sampling with local density-guided optimization, Rosetta-based all atom refinement and real-space B-factor calculations. Thank to Lux and Vadim. Great collaboration with Frank. Try it out!
doi.org/10.1038/s41596-022-00757-9
Angew Chem Int Ed Engl (2022) - Antibiotic with superior activity
.... CryoEM structure of a novel antibiotic derivative with enhanced antibiotic activity. Acts against Gram-negative bacteria by binding to the BAM complex, which integrates membrane proteins into the outer membrans. Great team effort by Carsten, Christoph and Biao to run this collaboration with the Mueller-lab from Helmholtz, Saarbruecken, Germany.
doi:10.1002/anie.202214094
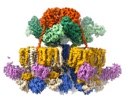
Nature (2022) - Resolving the AAA+ ATPase Puzzle in Slow Motion
.... The first high-resolution structure of the RuvAB complex on Holliday junctions reveals the chemomechanical coupling of ATP hydrolysis and mechanic work (DNA transport). In this study we use cutting-edge cryoEM, time-resolved studies and slower burning fuels to elucidate the entire nucleotide (hydrolysis and exchange) cycle. We reveal an internal signaling cascade within AAA+ ATPase hexamers that relays information between physically separated subunits to cycle between nucleotide exchange and hydrolysis. Our study provides a unifying model for the debated sequential and stochastic models in AAA+ ATPases. Heroic team effort especially from PDs Jiri and Dirk with support from a great team of people and discussion partners. (https://rdcu.be/cUgH9)
doi:10.1038/s41586-022-05121-1
RuvAB-HJ on Twitter: @MarlovitsLab
Current Opinion in Structural Biology (2022)
.... Review about Type-3 Secretion Systems resolved by cryoEM including novel insights about the structure of active T3SS. Thanks Julien (Bergeron, Kings College, UK) for this nice jointly work.
doi: 10.1016/j.sbi.2022.102403
Science Advances (2021)
.... Structures and structural states of human peptide transporters (PepT1 and PepT2) that also transport a variety of drugs revealed. Fantastic collaborative team effort with the Loew Lab (EMBL-Hamburg). Thanks Maxim and Jirka.
doi 10.1126/sciadv.abk3259
Nature (2021)
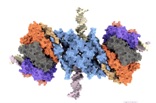
.... The first high-resolution structure of the ESX-5 Type-7 Secretion System from Mycobacterium tuberculosis critical for bacterial physiology and virulence. In this study we also show how the essential, yet enigmatic protein MycP stabilizes the entire complex, the effects to the structure in the absence of MycP, and dynamics of the cytoplasmic domains. Heroic team effort, especially from Catalin, Dirk, and Jirka and a wonderful collaboration with the Bitter and Houben labs in Amsterdam.
doi 10.1038/s41467-021-21143-1
“Type-7” trailer on Twitter: CLICK
Nature Communications (2021)
.... T3SS in action and at atomic resolution. This work shows the mechanism of gating of the T3SS to allow non-globular proteins (effectors) to be transported across several membranes. Moreover, Sean, Dirk, Niki und Jirka discovered that lipids are integral part of the T3SS and presented the ‘structure’ of an unfolded protein.
doi 10.1038/s41467-021-21143-1
“Type-3 in Action” trailer on Twitter: CLICK
Protein Science (2020)
.... Tools: Vadim presents a quantitative description to determine Gibbs free energy of unfolding as a proxy to assess protein stability in a reaction buffer. He developed MoltenProt to find stabilizing conditions for proteins (including membrane proteins) based on thermal unfolding measurements using changes of intrinsic fluorescence. We are grateful for wonderful collaborations with Susan Buchanan’s (NIH), Joao Morais-Cabral ‘s(Porto), Stephan Nussberger’s (Stuttgart), Maria Garcia-Alai’s and Christian Löw’s (both EMBL/Hamburg), and Kristina Djinovic-Carugo’s (Univ. Vienna) lab.
doi:10.1002/pro.3986 (online ahead of print)
Textbook: Molecular Biology of the Cell (Alberts et al)
.... T3SS: Oliver, the first graduate student in our lab, took images of isolated Type III secretion systems. These are now printed in chapter 23 (“Pathogens and Infection”) in the famous textbook Molecular Biology of the Cell (Alberts et al).
PNAS (2020)
.... Mechansims of F-actin bundlers (alpha-actinins): The actin cytoskeletal is a dynamic network of actin filaments and F-actin binding proteins such as alpha-actinins. In a collaborative work with the Djinovic-Carugo lab we show how Calcium modulated domain flexibility of alpha-actinins thus regulate actin bundling. Luciano was leading the electron microscopy part of that integrative structural study. Great work!
In collaboration with the Djinovic-Carugo (U Vienna/MFPL) lab.
doi: 10.1073/pnas.1917269117
Communications Biology (2020)
.... T3SS: Bacteria Exploited for Therapeutic Applications. Will we be harnessing the protein translocation power of the T3SS to treat various diseases in the future? This work demonstrates how such a system can be designed. In a collaborative effort, Antoine (graduate student in our lab and now post-doc in the Penninger lab) developed the basic principles for reprogramming the secretion system to deliver therapeutic molecules directly into cells. Fantastic!
In collaboration with the Penninger-Lab (IMBA/Vienna, LSI/Vancouver), Plückthun-Lab (U. Zürich), and Rabbits-Lab (Oxford)
10.1038/s42003-020-1072-4
Curr Top Microb Immunol. (2020)
.... T3SS: Book chapter in Current Topics in Microbiology and Immunology wtihin the series “Bacterial Type III Secretion Systems”: The structure of the Type III Secretion System Needle Complex - by Sean and Niki
doi: 10.1007/82_2019_178
BioRxiv (2019)
.... T3SS: Elucidation of the complete assembly pathway of the T3SS needle complex at 2.5 Angstroem of resolution - “Structural control for the coordinated assembly into functional pathogenic type-3 secretion systems”. Fantastic work from Nikolaus, Vadim, and Matthias, including many others in the lab, who contributed to fully understand the assembly process for the first time at unprecedented high resolution|
In collaboration with the DiMaio (Seattle), Wagner (Tübingen), Lea (Oxford) Labs
PNAS (2019)
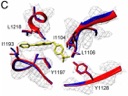
.... Human rhinovirus: Identification of an unexpected binding site for the antiviral drug OBR-5-340 determined by cryoEM. Great work from Jirka in the lab, who uncovered the site! In collaboration with Kirchmair (Bergen), Blaas (Vienna), Schmidtke (Jena) Labs
doi: 10.1073/pnas.1904732116
SciRep (2019)
.... Technology: High-throughput screening for detergent-solubilized membrane proteins. Congratulations to Vadim in the lab!
In collaboration with Garcia-Ali (Hamburg)
doi: 10.1038/s41598-019-46686-8
PloSBiology (2019)
.... T3SS: The type-3 secretion system needle filament is structurally polymorph.
In collaboration with the J. Galan (Yale) and A Loquet (Bordeaux) labs.
doi: 10.1371/journal.pbio.3000351
Nature Microbiology (2017)
.... Tuberculosis: Structure of the type VII secretion system from Mycobacteria elucidated for the first time. The high flexibility of specific domains suggests an as yet unseen mode of substrate recognition. Congratulations to Luciano in the lab, who solved the structure of this highly challenging sample.
In collaboration with the Bittner/Houben (VU Amsterdam) and Wilmanns (EMBL Hamburg) labs.
doi: 10.1038/nmicrobiol.2017.47
Nature communications (2017)
.... T3SS: Complete dissection of the regulatory elements within one of the most complex systems known (T3SS) to date, and application of basic conceptual principles and tools from synthetic biology to refactor and build a complete functional T3SS.
In collaboration with the Voigt lab (MIT/Boston).
doi: 10.1038/ncomms14737
PLoS Pathogens (2016)
.... T3SS: Biochemical, native mass spec and EM-characterization of the export apparatus of the T3SS.
In collaboration with the Robinson (Oxford), Galan (Yale) and Wagner (Tübingen) labs.
doi: 10.1371/journal.ppat.1006071
Nature methods (2015)
.... Technology: Novel methods paper for accurate atomic structure determination from density maps.
In collaboration with the Baker Lab (Seattle)
doi 10.1038/nmeth.3286
Annual Review in Microbiology (2014)
.... review “Bacterial type III secretion systems: specialized nanomachines for protein delivery into target cells”
doi: 10.1146/annurev-micro-092412-155725
Journal of Cell Biology (2014)
.... kinetochore paper in JCB.
In collaboration with the Westermann lab (Vienna)
Nature Structural and Molecular Biology (2014)
.... T3SS: type III secretion systems in action - in vitro and in situ - first demonstration and visualization of unfolded protein transport across membranes. Congratulations to Julia and Lisa in the lab for their heroic efforts!
doi: 10.1038/nsmb.2722
—jobs
.... accepting applications for several experts in:
[molecular biology and/or protein biochemistry] or [electron microscopy] (PostDoc, PhD, M.sc, TA-level)

StarMap